Plugins/Blocktype/Jmol
From Mahara Wiki
< Plugins
Jmol/JSmol plugin
Mahara artefact file blocktype jmol/jsmol plugin
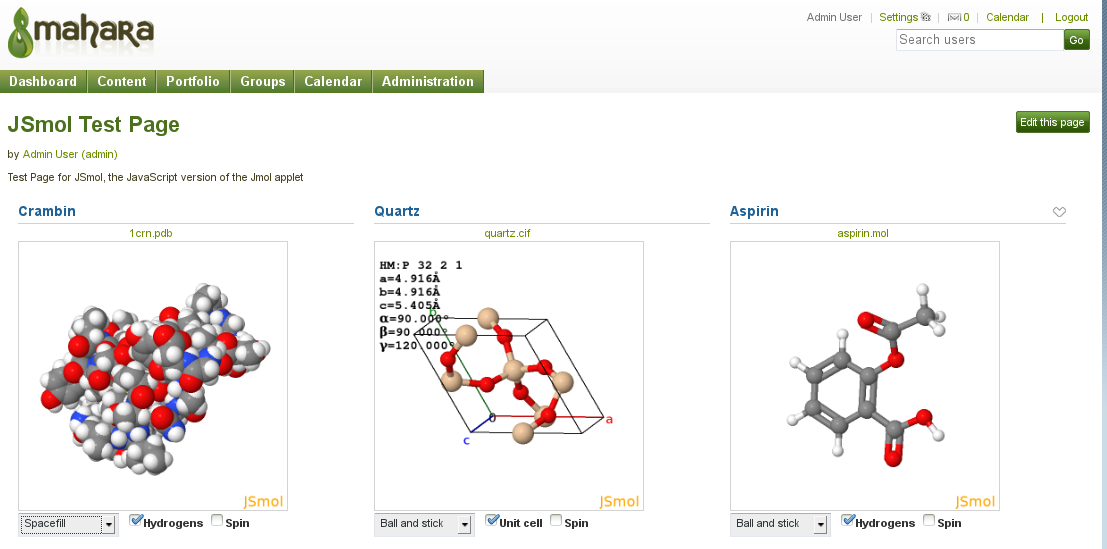
This plugin provides the interactive 3D display of chemical structure files, uploaded to Mahara, using Jmol/JSmol.
It is similar to other Mahara artefact file blocktype plugins such as the Embedded media plugin. Jmol/JSmol can use a variety of chemical structure file formats (chemical MIME types).
http://wiki.jmol.org/index.php/File_formats http://www.ch.ic.ac.uk/chemime/
This plugin currently uses the following subset of chemical structure file (chemical MIME) types, though others can be easily added (see below)
alc chemical/x-alchemy Alchemy cif chemical/x-cif Crystallographic Information File cml chemical/x-cml Chemical Markup Language hin chemical/x-hin HyperChem Input file mcif chemical/x-mmcif MacroMolecular Crystallographic Information File mol chemical/x-mdl-molfile MDL Molfile mol2 chemical/x-mol2 SYBYL Tripos Mol2 file format pdb chemical/x-pdb Protein Data Bank pdb.gz Gzipped Protein Data Bank pse chemical/x-pse PyMOL Session sdf chemical/x-mdl-sdfile Structure Data File xyz chemical/x-xyz XYZ Coordinate Animation format
Use
When editing the contents of a Mahara Page (View), drag the Jmol icon from the 'Files, images and videos' tab into your Page (View)
Then upload/select the appropriate file and set the appropriate applet width and height (default 300 px v 300 px)
For more advanced Jmol use, there are a couple of textarea controls that allow you to:
1. Specify an optional Jmol startup script to customise initial display of the applet http://chemapps.stolaf.edu/jmol/docs/
2. Enter optional Jmol.js JavaScript commands to add custom controls below the applet http://jmol.sourceforge.net/jslibrary/
Default script values are applied for general use.
NB Development has focused on functionality, and there may be security implications in running these scripts.
Installation
The latest version of the plugin, for Mahara 1.2, 1.3, 1.4, 1.5, 1.6 and 1.7 can be downloaded from Github
https://github.com/geoffrowland/mahara-artefact_file_blocktype_jmol
On unpacking, the /jmol folder (and its contents) is copied into the /artefact/file/blocktype folder of your Mahara installation.
Some other edits of Mahara files are required (or you can copy across the appropriate files from the download bundle).
To
/artefact/file/lang/en.utf8/artefact.file.php
add, immediately after the other file types
// Chemical $string['alc'] = 'Alchemy File'; $string['cif'] = 'Crystallographic Information File'; $string['cml'] = 'Chemical Markup language'; $string['hin'] = 'HyperChem Input File'; $string['mcif'] = 'MacroMolecular Crystallographic Information File'; $string['mol'] = 'MDL Molfile'; $string['mol2'] = 'SYBYL Tripos Mol2 File'; $string['pdb'] = 'Protein Data Bank'; $string['sdf'] = 'MDL Structure Data File'; $string['xyz'] = 'XYZ Coordinate Animation Format';
Then you need to edit /artefact/file/lib.php to 'force' the appropriate file mime-types. The precise details differ depending on the Mahara version
For Mahara 1.6: Immediately after
if (empty($data->filetype) || $data->filetype == 'application/octet-stream') {
$data->filetype = $data->guess;
}
add
// The browser may have been wrong, so use file extension to force some mime-types.////
$ext = $data->oldextension;
switch ($ext) {
case 'mm': $data->filetype = 'application/x-freemind';
break;
case 'alc': $data->filetype = 'chemical/x-alchemy';
break;
case 'cif': $data->filetype = 'chemical/x-cif';
break;
case 'cml': $data->filetype = 'chemical/x-cml';
break;
case 'hin': $data->filetype = 'chemical/x-hin';
break;
case 'mcif': $data->filetype = 'chemical/x-mmcif';
break;
case 'mol': $data->filetype = 'chemical/x-mdl-molfile';
break;
case 'mol2': $data->filetype = 'chemical/x-mol2';
break;
case 'pdb': $data->filetype = 'chemical/x-pdb';
break;
case 'sdf': $data->filetype = 'chemical/x-mdl-sdfile';
break;
case 'xyz': $data->filetype = 'chemical/x-xyz';
break;
}
For Mahara 1.4: Immediately after
if ($um->file['type'] == 'application/octet-stream') {
// the browser wasn't sure, so use file_mime_type to guess
require_once('file.php');
$data->filetype = file_mime_type($um->file['tmp_name']);
}
else {
$data->filetype = $um->file['type'];
}
add
// The browser may have been wrong, so use file extension to force some mime-types.////
require_once('file.php');
switch ($um->original_filename_extension()) {
case 'mm': $data->filetype = 'application/x-freemind';
break;
case 'alc': $data->filetype = 'chemical/x-alchemy';
break;
case 'cif': $data->filetype = 'chemical/x-cif';
break;
case 'cml': $data->filetype = 'chemical/x-cml';
break;
case 'hin': $data->filetype = 'chemical/x-hin';
break;
case 'mcif': $data->filetype = 'chemical/x-mmcif';
break;
case 'mol': $data->filetype = 'chemical/x-mdl-molfile';
break;
case 'mol2': $data->filetype = 'chemical/x-mol2';
break;
case 'pdb': $data->filetype = 'chemical/x-pdb';
break;
case 'sdf': $data->filetype = 'chemical/x-mdl-sdfile';
break;
case 'xyz': $data->filetype = 'chemical/x-xyz';
break;
}
To
/artefact/file/filetypes.xml
just before
<!-- General types -->
add
<!-- Chemical formats -->
<filetype>
<description>alc</description>
<mimetypes>
<mimetype>chemical/x-alchemy</mimetype>
</mimetypes>
</filetype>
<filetype>
<description>cif</description>
<mimetypes>
<mimetype>chemical/x-cif</mimetype>
</mimetypes>
</filetype>
<filetype>
<description>cml</description>
<mimetypes>
<mimetype>chemical/x-cml</mimetype>
</mimetypes>
</filetype>
<filetype>
<description>hin</description>
<mimetypes>
<mimetype>chemical/x-hin</mimetype>
</mimetypes>
</filetype>
<filetype>
<description>mcif</description>
<mimetypes>
<mimetype>chemical/x-mmcif</mimetype>
</mimetypes>
</filetype>
<filetype>
<description>mol</description>
<mimetypes>
<mimetype>chemical/x-mdl-molfile</mimetype>
</mimetypes>
</filetype>
<filetype>
<description>mol2</description>
<mimetypes>
<mimetype>chemical/x-mol2</mimetype>
</mimetypes>
</filetype>
<filetype>
<description>pdb</description>
<mimetypes>
<mimetype>chemical/x-pdb</mimetype>
</mimetypes>
</filetype>
<filetype>
<description>sdf</description>
<mimetypes>
<mimetype>chemical/x-mdl-sdfile</mimetype>
</mimetypes>
</filetype>
<filetype>
<description>xyz</description>
<mimetypes>
<mimetype>chemical/x-xyz</mimetype>
</mimetypes>
</filetype>
<filetype>
<description>mm</description>
<mimetypes>
<mimetype>application/x-freemind</mimetype>
</mimetypes>
</filetype>
You will have to edit /artefact/file/version.php to increase the value of $config->version (e.g. +1). Thin visit the administration interface to force the an update and load the chemical mimetypes into the Mahara database
This plugin includes a recent Jmol/JSmol version (13.1.16_dev) located in the /jsmol folder. However, JSmol code is frequently updated and you may wish to replace/update the contents of the /jsmol folder with the latest version, unpacked from http://chemapps.stolaf.edu/jmol/jsmol.zip
Finally, visit your Site Administration > Plugin Administration page to enable the plugin. Remember to select the chemical file types you wish to use from the configuration menu.
For the JSmol version, you need an HTML5-capable JavaScript-enabled browser. Recent versions of Firefox, Chrome, Safari, Opera and Internet Explorer 9 or 10 should work. IE 8 defaults to the Jmol Java Applet (you will need an up-to-date Java installation on the client computer. http://www.java.com).
During configuration and initial display of Jmol/JSmol embedded in a page, you may have to refresh the browser.
Remember to set appropriate server permissions for the new folders and contents.
Finally, visit your Site Administration > Plugin Administration page to enable the plugin. Remember to select the chemical file types you wish to use from the configuration menu file/jmol[config].
Of course, for the Jmol plugin, you will need an up-to-date Java installation on the client computer.
During configuration and initial display of the Jmol applet in a view, you may have to refresh the browser to see the applet embedded in the Page (View)
See Also
Jmol NIH Plugin
Chem 2D NIH Plugin
To Do
Currently, the plugin adds a default set of controls appropriate for basic organic chemistry
Add option buttons to the file-picker/edit interface for:
- default controls for biological macromolecules (e.g. .pdb files from PDB)
- default controls for crystal display filling unit cell(s) (e.g. .cif files from AMS or COD)
- other? reaction animations? orbital display? spectral display? virtual model kit?