Plugins/Blocktype/JmolNIH
From Mahara Wiki
< Plugins
Jmol NIH plugin
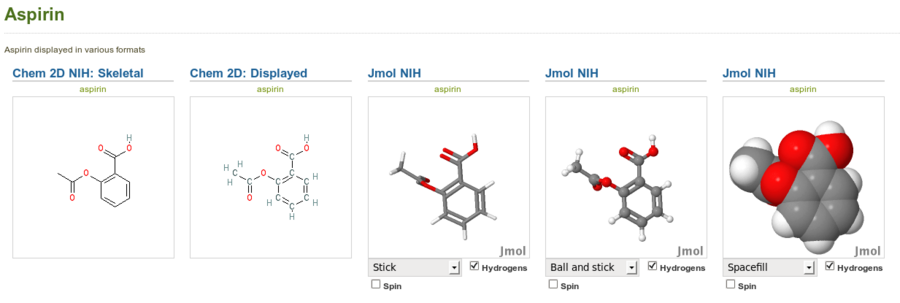
Aspirin displayed in stick, ball and stick and spacefill modes with the Jmol NIH plugin (right) alongside the same structure displayed with the Chem 2D NIH plugin (left). Note that this is just an image of the Jmol applets, not the interactive applets themselves.
This plugin allows 'instant' visualisation of interactive 3D chemical structures in Mahara.
It relies on the remarkable Chemical Identifier Resolver service developed by the Computer-Aided Drug Design (CADD) Group of the Chemical Biology Laboratory (CBL), National Cancer Institute (NCI), National Institute of Health (NIH) located at NCI-Frederick.
http://cactus.nci.nih.gov/chemical/structure
The Jmol NIH (jmolnih) plugin uses the Chemical Identifier Resolver service to return a 3D Structure Data File (sdf) for any meaningful chemical search term. Valid search terms include a trivial chemical name, systematic (e.g. IUPAC) chemical name, SMILES, InChI or InChIKey. The returned file is then rendered by the awesome Jmol Java applet. You can set the height and width of the embedded applet.
To avoid security issues with remote file access by the unsigned Java applet, the file contents are read via PHP into a string variable which is then loaded by Jmol. Ugly, but it works!
Use
When editing the contents of a View, drag the Jmol NIH icon (below) from the 'External feeds' tab into your View
Then enter in an appropriate search term (e.g. ethanol, morphine, buckyball, Cl/C=C\Cl) to get an (almost) instant 3D structure.
For more advanced Jmol use, there are a couple of textarea controls that allow you to:
1. Specify an optional Jmol startup script to customise initial display of the applet http://chemapps.stolaf.edu/jmol/docs/
2. Enter optional Jmol.js JavaScript commands to add custom controls below the applet http://jmol.sourceforge.net/jslibrary/
Default script values are applied for general use
Note that development has focused on the functionality of this, and there may be security implications of running these scripts. Happy to receive advice and guidance from Mahara security gurus about this.
Installation
The latest version of the plugin, for Mahara 1.2 and 1.3, can be downloaded from Github
https://github.com/geoffrowland/mahara-blocktype_jmolnih
On unpacking, the /jmolnih folders (and their contents) are simply copied into the /blocktype folder of your Mahara installation.
Remember to set appropriate server permissions for the new /blocktype/jmolnih folder and contents.
For the /jmolnih plugin you also need to download and unpack the latest Jmol stable version (currently 12.0.43), rename the unpacked folder (e.g. /jmol-12.0.43) to /jmol and copy this (and its contents) into the /lib folder of your Mahara installation to give /lib/jmol. Not all the contents of the jmol folder are actually required. However, Jmol code is frequently updated so it is convenient to just replace the /jmol folder with the latest version.
Finally, visit your Site Administration > Plugin Administration page to enable the plugins.
Of course, for the /jmolnih plugin, you will need an up-to-date Java installation on the client computer. http://www.java.com
During configuration and initial display of the Jmol applet in a view, you may have to refresh the browser.
See Also
Chem 2D NIH Plugin - to display images of 2D skeletal or full displayed structural formulae from NIH CADD Chemical Identifier Resolver service
Jmol Plugin Plugin - to provide interactive 3D display of chemical structure files in your Mahara Files repository
Todo
- Improve 'fallover' handling when NIH NCI/CADD Chemical Identifier Resolver service is not available. It is still in beta and so is occasionally off line for development.
- Ping NIH NCI/CADD server?
- Consider using cURL rather than PHP file_get_contents()?
- Perhaps some form of local caching of sdf data?
- Might also be solved if the Chemical Identifier Resolver is mirrored.
- In the near future, we intend to release some variants of the Jmol plugin to display local structure files (pdb, mol, cif, xyz, sdf cml etc) and directly display biological macromolecules from the RSCB Protein Data Base (PDB) service and mineral crystal structures from the Crystallography Open Database (COD)